

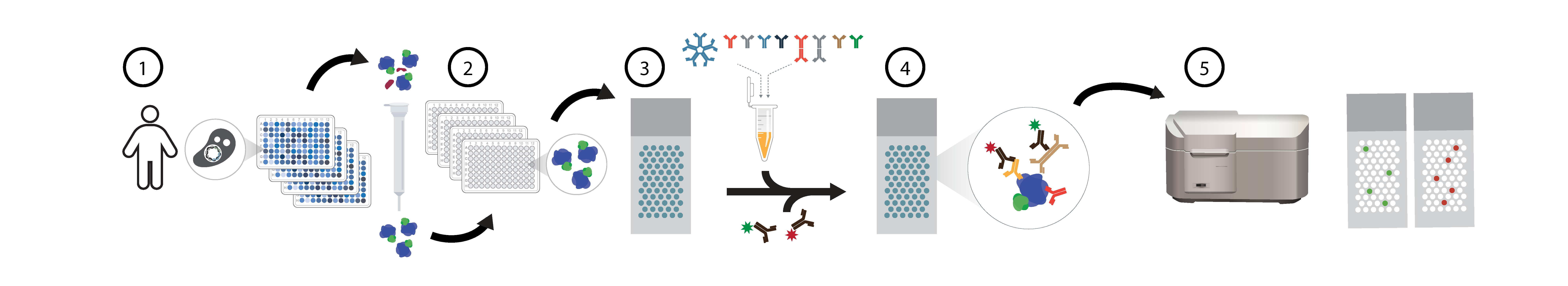
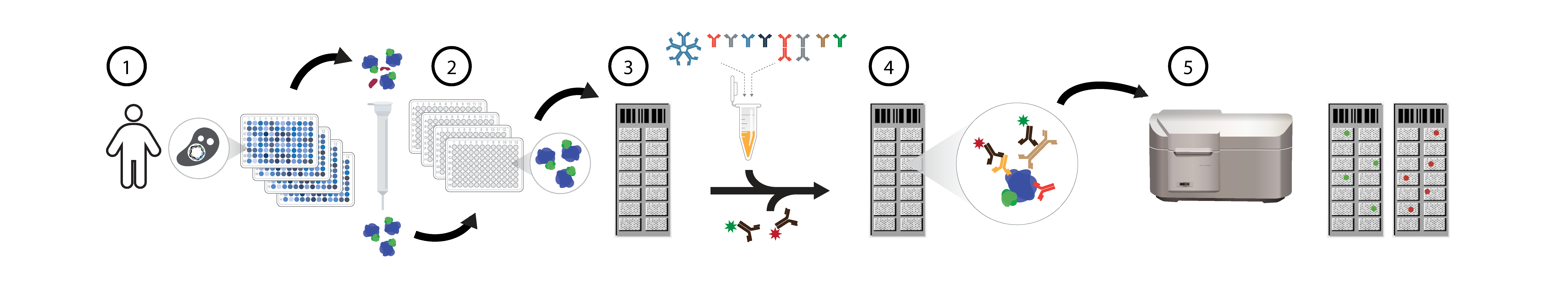
Overview of HuProt human proteome microarray seromics assays. For human proteome (HuProt) protein microarray assays, individual yeast clones are grown and 1) proteins purified via GST-tags, >21,000 unique human proteins are individually 2) printed in duplicate on nitrocelluose slides, reacted with patient serum samples 3), and dual stained with fluorescent 4) anti-IgG (635nm red) and anti-IgA (532nm green) secondary antibodies. Arrays are scanned 5) and data extracted using a Genepix microarrray scanner.
Current Platform: HuProt arrays interrogate 21,000 full-length human proteins (~80% of the known proteome) for autoantibody binding via fluorescent immunoassays. Each protein clone is sequence-verified (Venkataraman, Nature Methods, 2018); proteins are expressed as GST-His6 fusions in yeast, purified, and printed in duplicate on nitrocellulose slides. Versus known positive and negative controls across 997 patient samples, HuProt demonstrates 99.0% sensitivity and 99.7% specificity with median 5.1% intra-array CV between technical replicates. Arrays remain sensitive to known clinical autoantigens at up to 1:10,000,000 serum dilution. Nearly identical performance across serum and various plasma tube types.
Advantages:
To Build — Full-length hotspot neoantigen proteins: The HuProt microarray contains >21,000 full-length wild-type proteins but lacks hotspot neoantigens (e.g., KRAS G12D). We will create site-specific mutant versions of the top 150 frequency neoantigens from existing HuProt clones, enabling paired wild-type/mutant comparisons for all public neoantigens occurring at >0.17% frequency in TCGA while maintaining experimental consistency through single-source GST-His yeast production. Additionally, we will source 100 validated-functional human cytokines to complement the yeast-expressed proteins, as N-terminal GST-tagged yeast systems are not always ideal for functional cytokine production despite their proven success in autoantibody discovery.
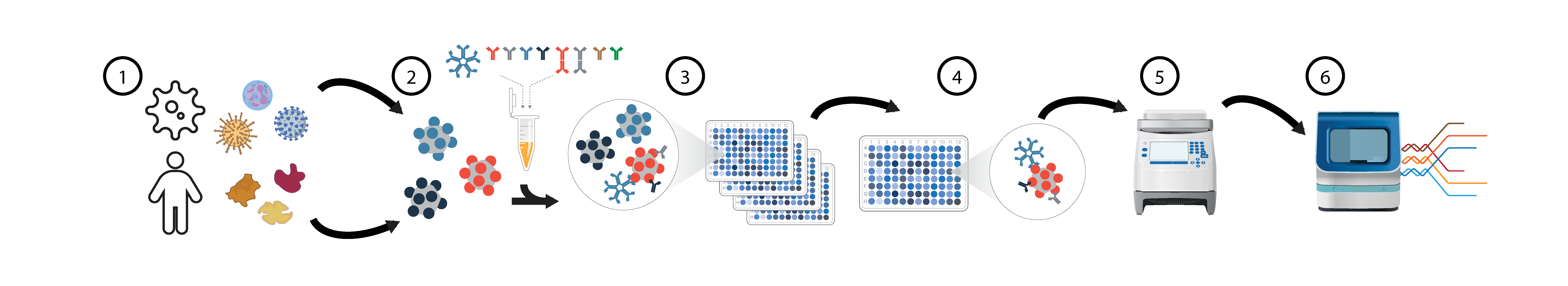
Overview of PhIP-Seq seromics assays (HuScan, VirScan, others). For phage display immunoprecipitation-sequencing (PhIP-Seq) serology assays (A) we utilize both a 1) >700,000 peptide 49mer human proteome library (HuScan) and a >480,000 peptide 62mer pan-viral library (VirScan). 2) Individual serum samples and controls are 3) reacted with each library and 4) antibody-bound phages immunoprecipitated with protein A/G beads. Individual samples are expanded, 5) given PCR barcodes, and pooled for 6) Illumina sequencing.
Current Platform: PhIP-Seq combines synthetic phage display libraries with immunoprecipitation and sequencing readout for massively multiplexed serology. Our plan combines existing libraries with new ones into a single run; this demonstration atlas utilizes HuScan (730K peptides) deveolped at UCSF/CZB and VirScan (480K peptides) developed at UCSF/CZB.
Advantages:
To Build — IEDB Pan-Exposome PhIP-Seq Library: We designed a pan-exposome PhIP-Seq library that includes >198,000 62-mer peptides from nearly 2,000 species, covering all known human-reactive antigens from the Immune Epitope Database (IEDB) assembled across >19,000 publications, plus potential human-reactive antigens from UniProtKB Swiss-Prot entries flagged as 'Biotechnology', 'Pharmaceutical', or 'Allergen'. This enables simultaneous epitope-level profiling of immune responses to viruses, commensal and pathogenic bacteria, vaccine antigens, allergens, foods, pet exposures, industrial proteins, processed-food enzymes, household cleaning products, and other xeno-exposures, generating a detailed "immune exposure history" for each individual.
To Build — Neoantigen Mutation-Site PhIP-Seq Library: We obtained tumor-specific mutations from Thorsson et al. (2018) covering 10,437 tumors across 33 TCGA cancer types, identifying 29,118 public neoantigens occurring in ≥3 patients via protein-coding missense SNVs or indels. For each mutation site, we designed a multi-window library of 33-mer peptides capturing LEFT/CENTER/RIGHT variants of both tumor and wild-type sequences (6 versions per site), with GGGS padding for N/C-terminal or premature-stop mutations and three alternative DNA encodings per peptide for synthesis success. The final neoantigen PhIP-Seq library contains 179,838 unique peptide-bearing phages.
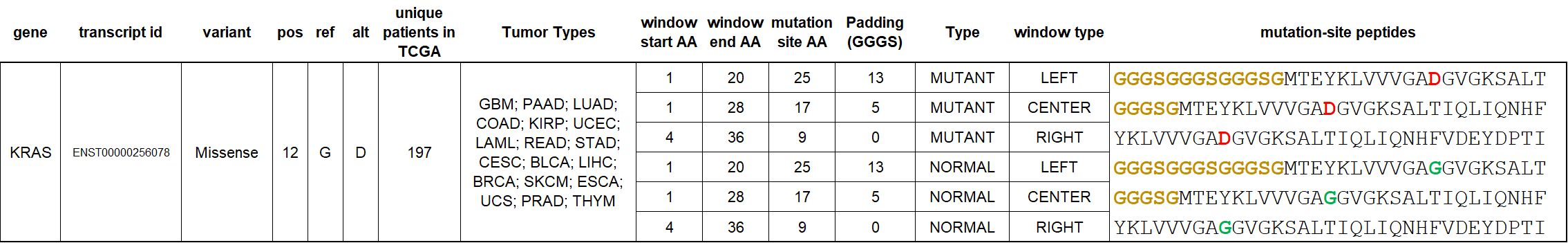
Public tumor neoantigen PhIP-Seq peptide design for the canonical KRAS G12D mutation occurring in 1.77% of all TCGA cancer patients. This and other library 33-mer peptides include triplicate windows of mutant and wild-type versions at each mutation-site; for terminal peptides such as KRAS G12D – flexible GGGS padding is used to preserve equal peptide lengths across the library. For all other draft KRAS library variants (CENTER versions only), see: https://caatlas.com/genes/KRAS.html
To Build — High-Confidence Dark Proteome PhIP-Seq Library: We obtained experimentally-supported alternative open reading frames from OpenProt v2.2, which predicts all possible ORFs ≥30 codons across the human transcriptome and validates them with 177 mass spectrometry datasets and 131 ribosome profiling experiments. We selected high-confidence "AltProts" (alternative proteins from unannotated ORFs with no sequence identity to canonical proteins) validated by ≥2 unique peptides, then designed overlapping 33-mer tiles with 17-AA overlaps covering full sequences, using GGGS padding for AltProts <33 amino acids. The final dark proteome PhIP-Seq library contains 383,685 unique peptide-bearing phages.
To Build — Human Peptide Hormone PhIP-Seq Library: Short signal peptides are not always adequately represented in PhIP-Seq libraries. Our draft hormone-focused PhIP-Seq library contains 6,520 unique phage-bearing peptides representing 356 entries (290 human peptide hormones and 66 peptide drugs) obtained from the HORDB database. Each peptide hormone is displayed in several redundantly windowed contexts (left, center, right, or repeated across the 34-mer display window) with 5 technical replicates per version.
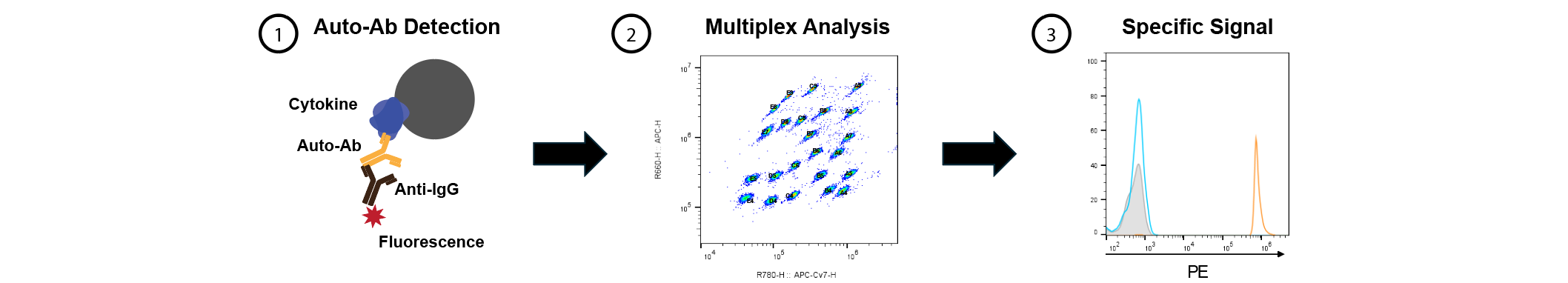
Overview of anti-cytokine autoantibody assays: Binding will be tested by multiplex particle-based assays for autoantibodies to known-functional human cytokines and chemokines.
Current Platform: The Paris team will conduct high-throughput multiplex IgG autoantibody testing using ELISA and cytokine multiplex panels, employing particle-based flow cytometry assays to screen for 46 different autoantibodies against cytokines involved in anti-tumor responses, immune regulation, and inflammatory pathways. These validated techniques require minimal sample volumes (microliters), are highly sensitive (SIMOA®), automated, and rapid, enabling large-scale detection of autoantibodies of multiple isotypes (IgG, IgM, IgA). Additionally, the team will assess the neutralizing activity of these autoantibodies against cytokines to demonstrate their functional impact.
Advantages:
To Build — Assays validating novel targets found in discovery phase: Additional interruption assays will be built to validate naturally-occurring checkpoint-like autoantibodies occuring in various populations with good or bad cancer outcomes.
Overview of HuProt gastketed minichip microarray seromics assays. For gasketed minichip protein microarray assays, discovery identified outlier yeast clones are grown and 1) proteins purified via GST-tags, either 384 or 768 individual proteins are 2) printed in duplicate on nitrocelluose slides along with independently sourced 3rd party proteins, reacted with patient serum samples 3), and dual stained with fluorescent 4) anti-IgG (635nm red) and anti-IgA (532nm green) secondary antibodies or other isotypes (IgG1, IgG2, IgG3, IgG4, IgA1, IgA2, IgM, IgE, IgD). Arrays are scanned 5) and data extracted using a Genepix microarrray scanner.
Current Platform: HuProt minichips utilize gaskets to efficiently interrogate protein subsets with similar performance to full-proteome HuProt arrays. HuProt minichips use identical protein preparations and nitrocellulose surfaces as full HuProt arrays; proteins can be optionally printed in dilution ladders for improved quantitation. Technical performance (CV, sensitivity, specificity) and data output comparable to full arrays.
Advantages: